Setting up Successful Purification Protocol with Ni-Penta Resins
May 07 2017 0 Comments Tags: Affinity purification, EDTA-resistant, HEK 293 cell protein purification, His-tagged protein purification, IMAC resins, Insect cell protein purification, Ni-Penta resins, Protein function, Protein structure, recombinant protein production
The purification conditions allowing for the maximal protein yield and purity may vary significantly depending on the characteristics of a given target protein. Finding the best conditions frequently require optimizing the key parameters through small-scale trial and error approaches.
1. Setting up binding conditions for target protein
In most successful purification protocols, the ratio for binding is 1 mL of resins for every 10 – 50 mL of eukaryotic culture supernatant; and 1 mL for 10 ~ 20 mL E.coli lysate.
To set up protocol to purify a new protein, it is recommended to start with the ratio of 1 mL resin for every 20 mL of culture supernatant or lysate. It also helps to estimate the amount of target protein in the sample and adjust it to approximately 0.5 mg/mL.
When working with an eukaryotic expression system that may contain unknown agents and factors interfering the binding of target protein, it is recommended to dilute the culture supernatant 1:1 or 1:2 with the Equilibration buffer (for example, 20 mM sodium phosphate, 0.5 M NaCl, pH 7.4).
If the target protein abundance is very low, enrichment of the target protein will help accelerate the binding equilibrium. Alternatively, allow binding for longer time with constant, gentle agitation (ie. on a rocking platform). If the protein is large (eg. >100 kDa), longer incubation time may also be necessary.
2. Determination of the optimal imidazole concentration in Washing and Elution.
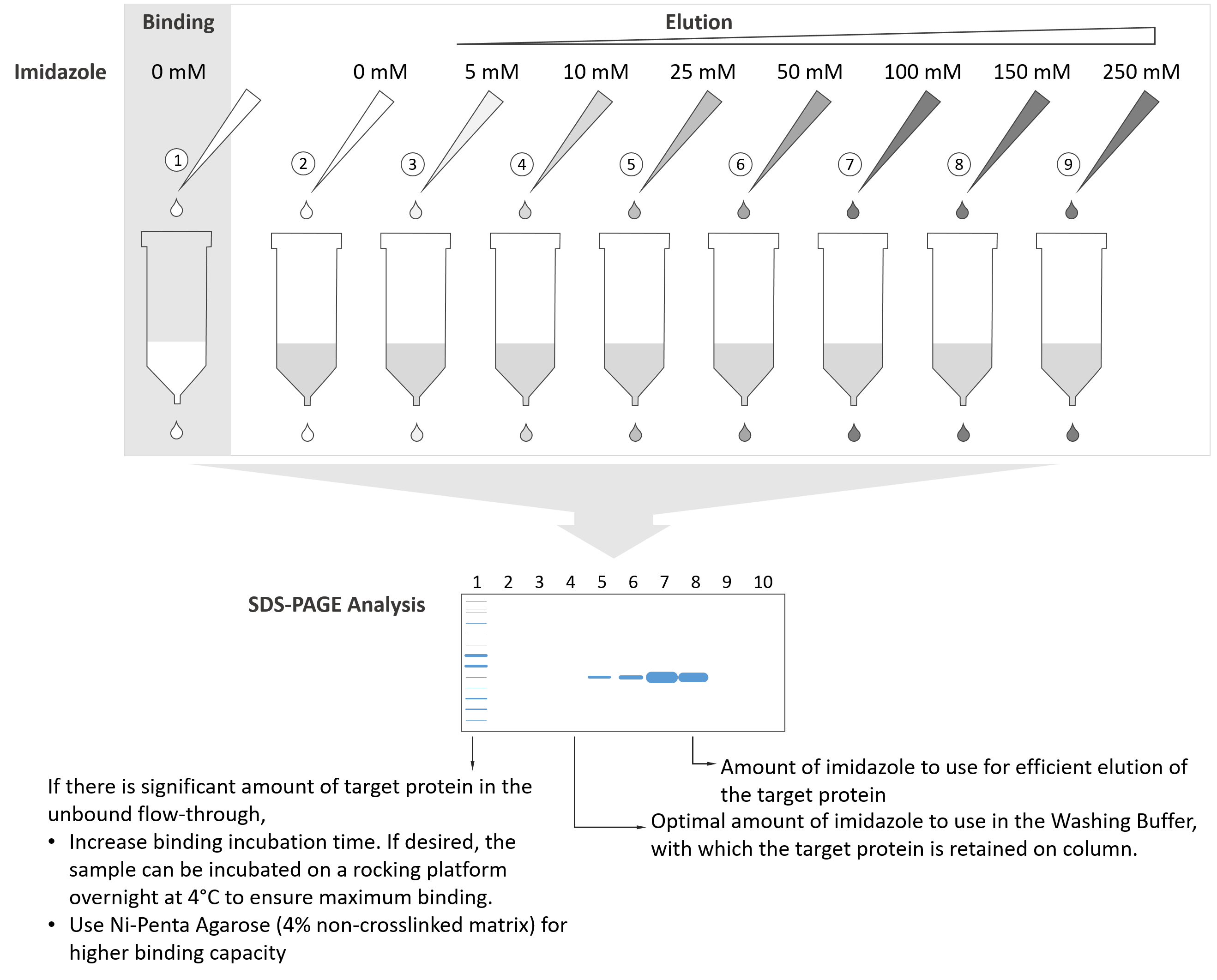
Do NOT add imidazole in the sample or the Equilibration buffer.
- Load the sample on the resins that have been equilibrated (for example, equilibrate with 5CV of Equilibration buffer), incubate and allow enough time for maximum binding.
- Collect the flow-through to determine the degree of binding.
- Then sequentially add a series solutions of increasing imidazole concentration (from 0 ~ 300 mM) to the column. For example, 0 mM, 5 mM, 20 mM, 50 mM, 100 mM, 150 mM, etc., 5 ~ 10 CV for each solution.
- Collect fractions from eluates and analyze them on SDS-PAGE.
3. Further optimization of the purification process
Once your target protein can be purified from Ni-Penta resins with decent yield and satisfactory purity, it may be desired to further optimize more parameters to achieve the best purification result. Here are some key parameters need to be optimized:
- Determine the optimal volume of resin required for the purification of a specific protein of interest. Optimal results will be obtained when the capacity of the resin matches the amount of target protein.
- Finding the optimal buffer composition: high salt concentration (for example, optimal purity can typically be achieved with buffer containing 300 mM NaCl at pH 8.0 for some proteins), pH, as well as addition of the right type of detergents at the right concentration (this is particularly important for membrane proteins or the proteins prone to aggregate).
- For automated applications, there are more parameters required to optimization, including:
- Sample loading flow rate
- Sample volume
- Imidazole gradients during wash
- Wash flow rate
- Wash volume
- temperature
0 comments

Leave a Comment